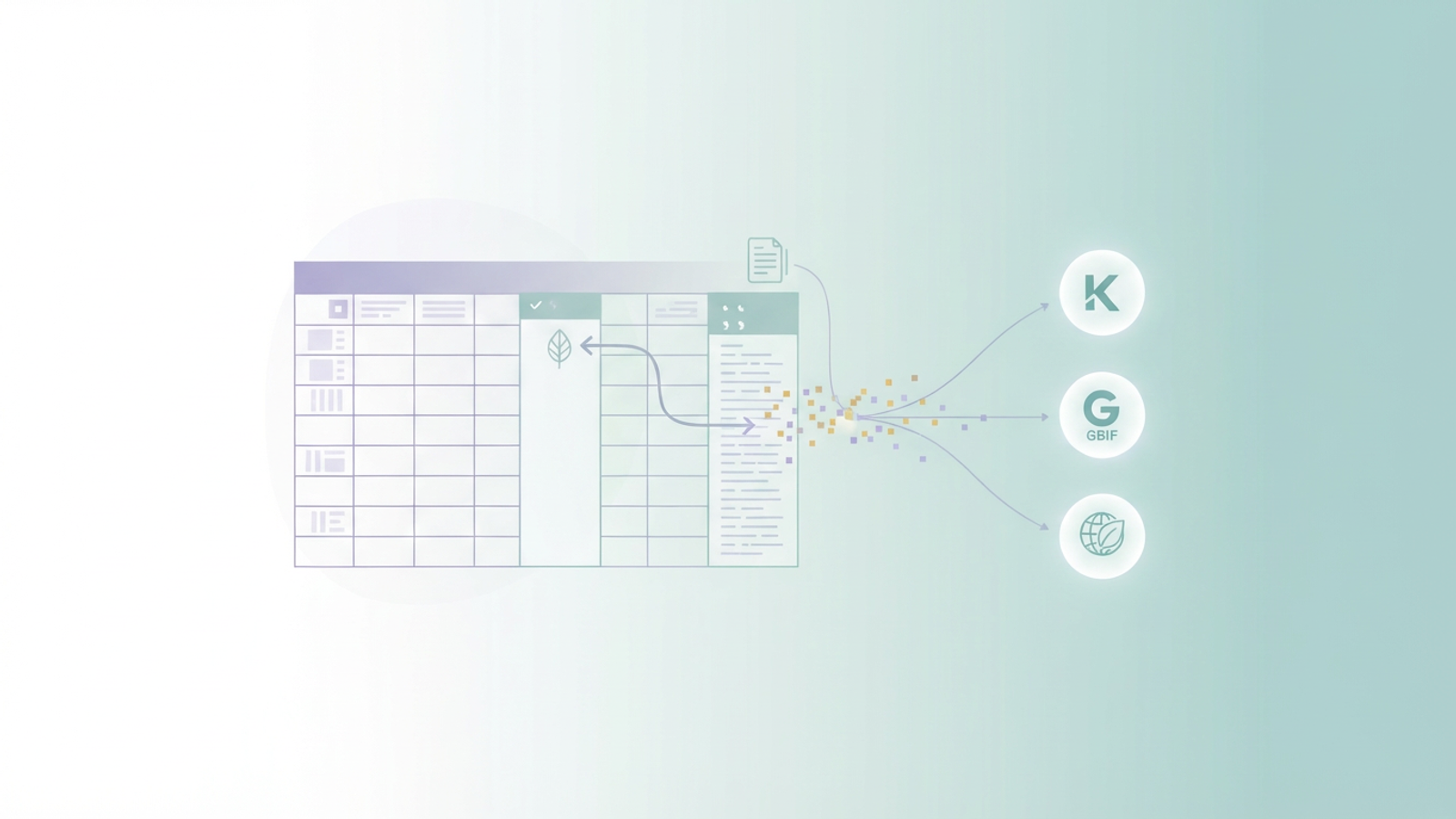
Taxonomy is a living science. A single plant species might have dozens of synonyms generated over centuries of study, reclassification, and historical revision. For the researcher, this presents a massive data challenge. You aren't just managing a list of plants; you are managing the history of their names.
At the heart of this system is botanical nomenclature, the formal scientific naming of plants. While the International Code of Botanical Nomenclature (often searched by its historical acronym ICBN, though now formally known as the ICN) provides the strict rules for how names are constructed and prioritized, it does not provide the tools to manage them.
Researchers, museum curators, and biodiversity data scientists often find themselves trapped in a manual workflow. They spend hours cross-referencing names against databases like Kew’s Plants of the World Online (POWO), GBIF, or World Flora Online to validate accepted names and compile synonyms. Traditional spreadsheets struggle to handle this. Manual entry leads to typos, and standard formulas cannot easily parse complex author citations.
This is where Quadratic offers a solution. By combining the familiarity of a spreadsheet with the power of Python, researchers can automate the retrieval and cleaning of taxonomic data, turning a week of manual curation into a reproducible, automated workflow.
The challenge of modern botanical nomenclature
Plant names change for many reasons. New phylogenetic studies based on DNA sequencing often reveal that what we thought were two distinct genera are actually one, or vice versa. When a name changes, the international rules of botanical nomenclature dictate how the new name is applied and what happens to the old one. This creates a constant need for "synonym management."
For a data scientist or botanist, this creates a specific, repetitive pain point often called the "copy-paste trap." The workflow usually looks like this: search a species name in a browser, copy the accepted name and its synonyms, paste them into a spreadsheet, and repeat for hundreds of rows. This process is slow, but more importantly, it is prone to human error. A misspelled specific epithet or a misplaced comma can break downstream data analysis.
Data hygiene is further complicated by author citations. In strict scientific contexts, a name is followed by the abbreviation of the author who named it (e.g., Helianthus annuus L.). However, for data analysis or ecological modeling, you often need just the clean binomial name. Inconsistent formatting—where one source uses "L." and another uses "Linnaeus"—makes merging datasets incredibly difficult.
Why standard spreadsheets fail at taxonomy
Most researchers default to standard spreadsheet software for data analysis because it is accessible, but these tools have significant limitations when dealing with biological data.
The first issue is static data. Once you paste a synonym list into a standard cell, it becomes "dead" data. It is disconnected from the source authority. If the accepted name changes next year due to a new revision in the code of botanical nomenclature, your spreadsheet remains outdated until you manually check it again.
The second issue is data cleaning. Stripping author citations from a name string is deceptively difficult using standard spreadsheet formulas. Logic that relies on finding the first space or comma is brittle; it breaks as soon as it encounters a subspecies or a variety. You often end up with a column of half-cleaned names that requires manual correction.
Quadratic bridges this gap. It provides a "middle ground" where you can use Python directly within the cells. You don't need to be a software developer or set up a complex local environment to use it. You simply write a script to fetch and clean the data, and the results populate your grid.
Tutorial: Automating synonym lookup with Python
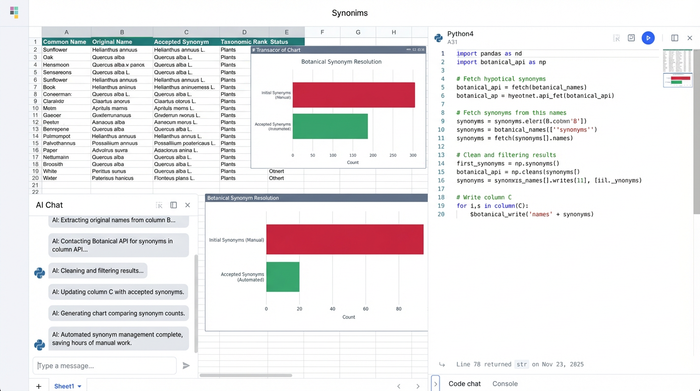
In this workflow, we look at how a researcher can move from a manual search process to an automated pipeline using Quadratic. The goal is to take a "dirty" list of species names and generate a clean, comma-separated list of synonyms and a column citing the source.
Step 1: Ingestion and batch lookup
Instead of searching for each species one by one, you can use Python in Quadratic to query authoritative APIs, such as GBIF or the Open Tree of Life, for your entire list at once.
In a standard workflow, you would have a column of input names. In an adjacent cell, you can write a Python script that references that column. The script sends the names to the API and requests the taxonomic backbone for each. The result is returned instantly into the spreadsheet. You input the list once, and the external database fills in the blanks.
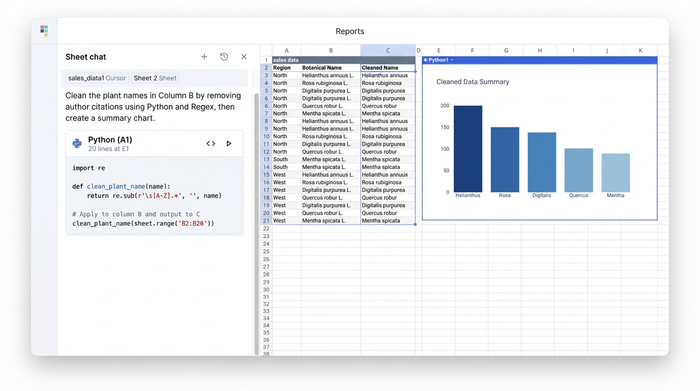
Step 2: Data cleaning and removing citations
A common requirement in botanical nomenclature definition and data curation is separating the name from the author. You might receive a string like "Helianthus annuus L." but you only need "Helianthus annuus" for your visualization software.
In Quadratic, you can handle this using Python’s string manipulation libraries or Regular Expressions (Regex). Rather than writing a nested formula that breaks easily, you write a short script that identifies the pattern of the author citation and removes it. The output is a dedicated 'Synonym List' column containing only the clean names, ready for analysis.
Step 3: Source validation
In scientific publishing, data must be traceable. If you list a synonym, you must state which authority claims it is a synonym. During the API query, you can instruct the script to pull the source URL or database name (e.g., "Kew POWO" or "GBIF Backbone Taxonomy") into a 'Botanical Literature Source' column.
This ensures that every row in your dataset has a direct lineage to a trusted authority, adhering to the rigor expected in the field.
Ensuring compliance with the code of botanical nomenclature
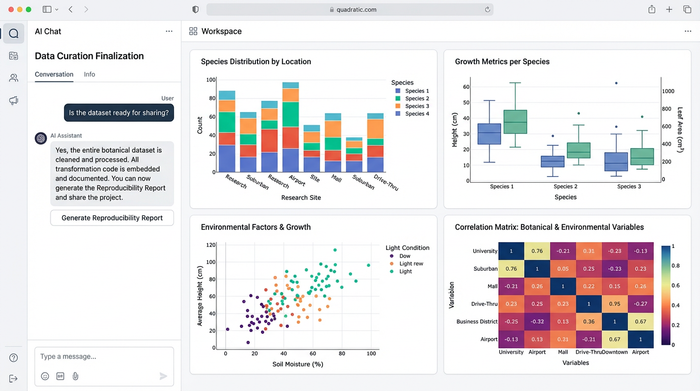
Accuracy is the currency of science. When you automate your workflow, you ensure that you are always using the most current names as defined by the botanical nomenclature code. You remove the risk of transcribing a name incorrectly or relying on an outdated field guide.
Furthermore, working in Quadratic makes your research reproducible. In the global context, where researchers might be searching for nomenclatura botánica resources in various languages, having a universal logic layer is essential. If a peer reviewer or an editor asks how you derived your list of synonyms, you don’t have to say "I searched for them." You can show them the code in the cell.
This transparency is critical when dealing with the international code of botanical nomenclature ICBN (or ICN). It proves that your data adheres to the accepted rules and that your methodology can be audited and repeated by others.
Conclusion: From manual lists to dynamic data
The shift from manual copy-pasting to automated curation changes the nature of botanical research. It frees taxonomists and data scientists from the drudgery of data entry, allowing them to focus on the biological implications of their work.
By using Quadratic, you respect the complexity of botanical nomenclature without being overwhelmed by the data management it requires. You gain the power of a developer’s toolkit within the interface of a data science spreadsheet, ensuring your data is clean, compliant, and ready for discovery.
If you are managing complex species lists and struggling with synonyms, try loading your data into Quadratic to experience how Python can streamline your workflow.
Use Quadratic to curate botanical nomenclature data
- Automate synonym lookup across entire species lists by querying authoritative botanical databases (like GBIF or POWO) directly from your spreadsheet, eliminating manual copy-pasting.
- Clean and standardize botanical names with Python directly in cells to effortlessly remove author citations and inconsistent formatting, preparing data for analysis and visualization.
- Ensure data accuracy and compliance by automatically capturing source URLs or database names for every synonym, providing traceability and adhering to the International Code of Botanical Nomenclature (ICN).
- Create reproducible taxonomic workflows by embedding Python scripts, allowing you to easily update your data as nomenclature evolves and share auditable methods with peers.
Ready to streamline your botanical data management? Try Quadratic.